virginie rougeron research evolution adaptation history population genetic
During the course of my carrier, I supervised several Master students, PhDs students, and Postdoc fellows.
As part of the research community, I actively participate in different research panels and instances
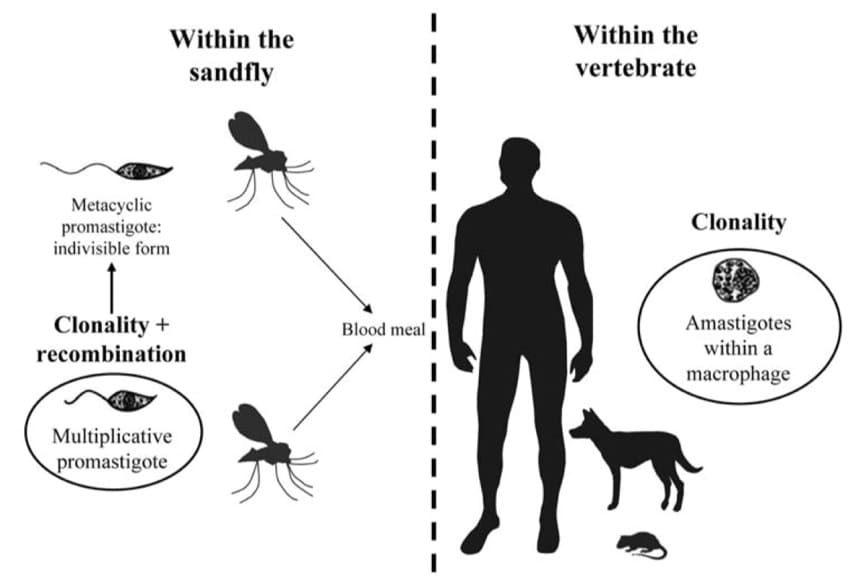
Mode of reproduction of Leishmania parasites
Resistance factors to Plasmodium
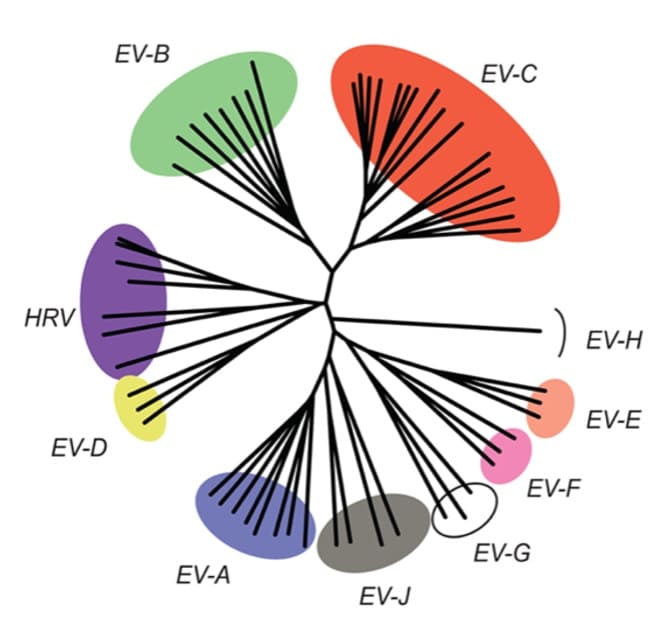
Evolution and adaptation of enteric viruses
Evolutionary history and adaptation Plasmodium parasites
Ecological adaptation to anthropogenic changes of chacma baboons
Welcome to my website
“I am a permanent staff of the CNRS since 2015, where I am developing multiple projects on the evolutionary and adaptive history of organisms. I have been working for six years in the Laboratory MiVEGEC first in Gabon, then in France, where my research was focusing on pathogens evolution. In 2020, the CNRS awarded me the bronze medal. During the course of my career, I have been the head of the team SHAPE and then HEAT in the laboratory MiVEGEC for 4 years.
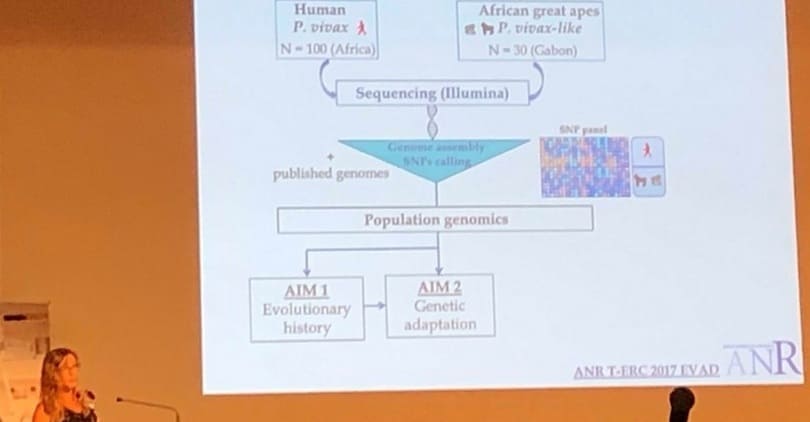
I have been the PI of several research funded projects, including one ANR-17-ERC3-002 on “Evolutionary adaptation of human Plasmodium vivax“, which lead to the sequencing of the first genome of African great apes P. vivax-like parasites. I have also been the PI of a PEPS ECOMOB project, which aimed to study the origin of the human P. vivax. This project leads to the identification of a southeast Asian origin of this parasite. Currently, I am the PI of an ANR JCJC that aims to study the evolutionary history and adaptation of pathogens in South America and of an annual CEBA project in which the main objective is to characterize non-human primate Plasmodium circulation in French Guyana.
Since December 2021, I moved to George in South Africa to join the International ‘Research Laboratory Reconciling Ecological and Human Adaptations for a Biosphere based Sustainability’, where I am implementing a large research project on the adaptation of chacma baboon populations in Austral Africa.”
Research
As a researcher in evolutionary biology, I have been for a long time specializing in the study of the evolutionary and adaptive history of organisms.
For 16 years now, I have been working on where, when, and how pathogens arose, adapted to new hosts, and evolved, which are fascinating questions in the world of infectious diseases. My interest in these questions was confirmed through my various research experiences, during which I was able to discover many facets of the evolutionary ecology of infectious diseases and carry out enriching field experiments. It is by combining these facets that we discovered a new species of parasites of the genus Plasmodium (agents of malaria), in African great apes, called Plasmodium vivax-like. Since this discovery, I have been developing my research projects around the evolutionary history and adaptation of this incredible parasite and other related pathogens.
Now that I integrated the International Research laboratory REHABS (‘Reconciling Ecological and Human Adaptations for a Biosphere based Sustainability’) based in George, South Africa, I am starting a new project on the evolutionary adaptation of chacma baboon populations to urban environments.
Supervision & Teaching
“During the course of my carrier, I supervised several Master students, PhDs students, and Postdoc fellows.”
I also participated in different Master Modules such as “Quantitative epidemiology”. As part of my new affiliation in REHABS, I will actively participate in the implementation of a Master module on “Conservation genetics and genomics”.
Do not hesitate to contact me directly by email for any interest and to be enrolled in our research team.
Membership in Panels
“As part of the research community, I actively participate in different research panels and instances”
MEEDIN – Centre for Research on the Ecology and Evolution of DiseaSes
Montpellier – link to the site
INEE Scientific Council – link to the site
Date Line
PhD
Postdoc 1
Postdoc 2
Evolutionary history and adaptation of Plasmodium parasites
Ecological adaptation to anthropogenic changes of chacma baboons.
PhD

Postdoc1
Postdoc2
Evolutionary history

Ecological adaptation
PhD
Postdoc 1
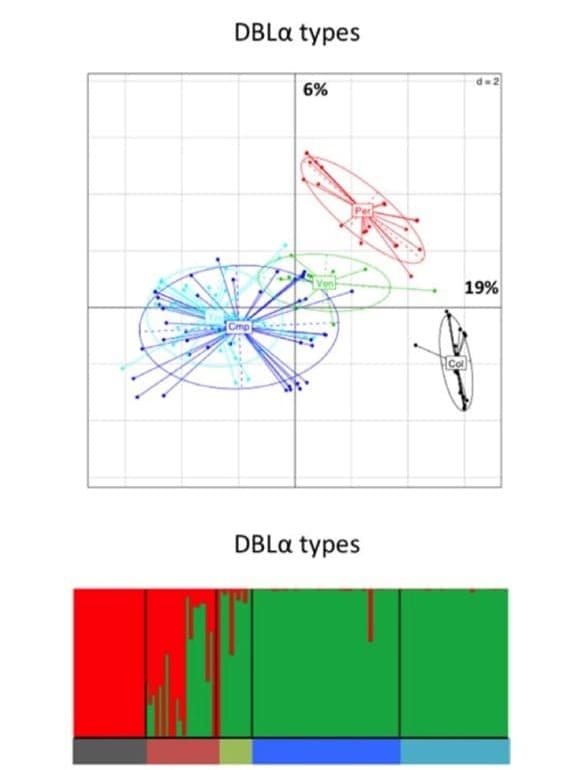
Genetic relationship between South American P. falciparum populations, based on var DBLα types. Principal component analysis, where the colored dots represent the population isolates, and the colored ellipses represent 95% of the genetic variation within each population. Percentages of inertia are displayed directly along the respective axes (first axis: horizontal; second axis: vertical). South American population structure inferred by Bayesian clustering. Each isolate is a column and is partitioned into K colored components (K = 2).
Plasmodium parasites
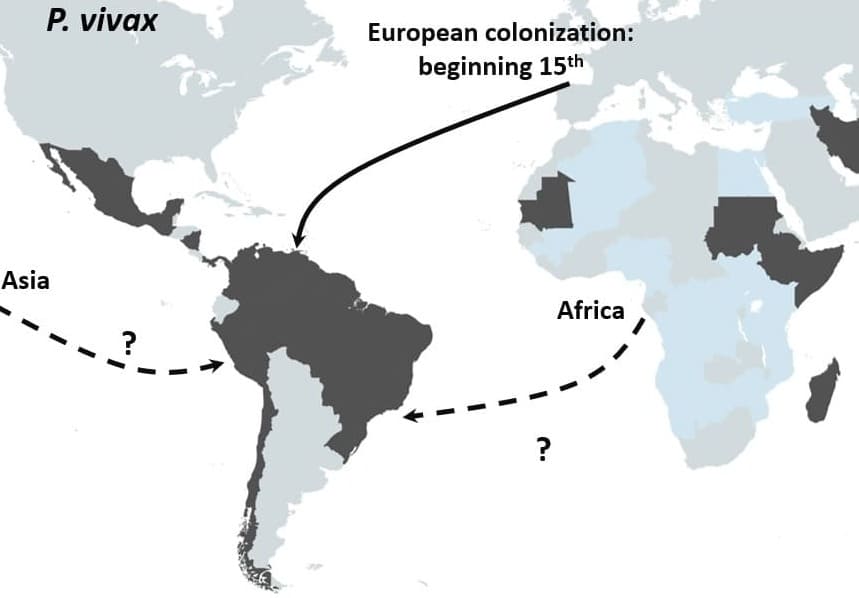
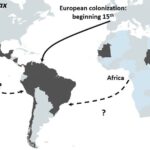
Schematic representation of the proposed origin in America of P. vivax. Main routes of the transatlantic slave trades, major European Empires between the 15th and 19th centuries in America are indicated with arrows. When the migration route and origin have been confirmed by genetic studies, arrows indicate the direction of the transfer. When migrations routes are still under debate, dashed arrows indicate the sense of the suggested transfer.
Field experience and teamwork
” After 16 years of research in ecology and evolution of organism’s adaptation, I am more than happy to collaborate, discuss, participate in conferences and workshops. My team has now high expertise in Plasmodium and more largely pathogens population genetics and genomics and is as such involved in multiple projects and formation.
We implemented multiple new protocols and technics to work mainly on non-invasive specimens. Finally, we also developed very good skills in samples collection in the field, going from non-human primate fecal samples, tsetse flies, mosquitoes, and other unexpected specimens. We are very happy to share our experience and help in any research project, do not hesitate to contact me for further information “