Research evolution adaptation plasmodium vivax-like african great apes humans
Infectious diseases represent the second cause of death worldwide
We propose to generate a radically global understanding of how pathogens adapt to new environments
study the pathogen evolutionary history the different P. vivax clades through the generation of the first P. vivax-like genomes
Completed Project
Recent News
Contact
- REHABS International Research Lab South Afrrica
- rougeron.virginie@gmail.com
- 06 46 64 23 18
More information about the project
Main Publications

@ V.Rougeron
Abstract
We propose to generate a radically global understanding of how pathogens adapt to new environments, using Plasmodia as model system, by integrating cutting-edge knowledge and technology in the field of genomics, population genetics, informatics and evolution. The malaria agent Plasmodium vivax, albeit less malignant than Plasmodium falciparum, is responsible for severe and incapacitating clinical symptoms with significant effects on human health. P. vivax is an ideal model because during its evolutionary history it has emerged in different hosts (i.e. chimpanzees/gorillas) and repeatedly colonized new human populations. It has thus shown ability to successfully adapt to different environments. The AIM of this proposal is to study the pathogen evolutionary history the different P. vivax clades through the generation of the first P. vivax-like genomes and of new African P. vivax genomes. ‘
Funding
ANR T-ERC EVAD 2017, “Evolutionary history and genetic adaptation of Plasmodium vivax”
Principal Investigator : Virginie Rougeron – 148,824€ ; 18 months ; December 2017 – May 2019.

ANR
Partners



“This project allowed first the sequencing of the first P. vivax-like genomes of African great apes. Then, our next study provides one of the most detailed views of the worldwide distribution of P. vivax population genetic diversity and demographic history. Our work investigated not only the phylogenetic relationship between P. vivax infecting human and apes but also different features of P. vivax genetic variation worldwide to test different hypotheses of an African or Asian origin. We showed that P. vivax is a sister group and not a sublineage of P. vivax-like. Genetic diversity in P. vivax-like is richer than in P. vivax, consistent with a strong bottleneck in this lineage that gave rise to P. vivax. Our results based on whole-genome sequencing data support an out-of-Asia origin, rather than an African origin, for the world populations of P. vivax, with a signal of stepping-stone colonization events accompanied by serial founder effects.”
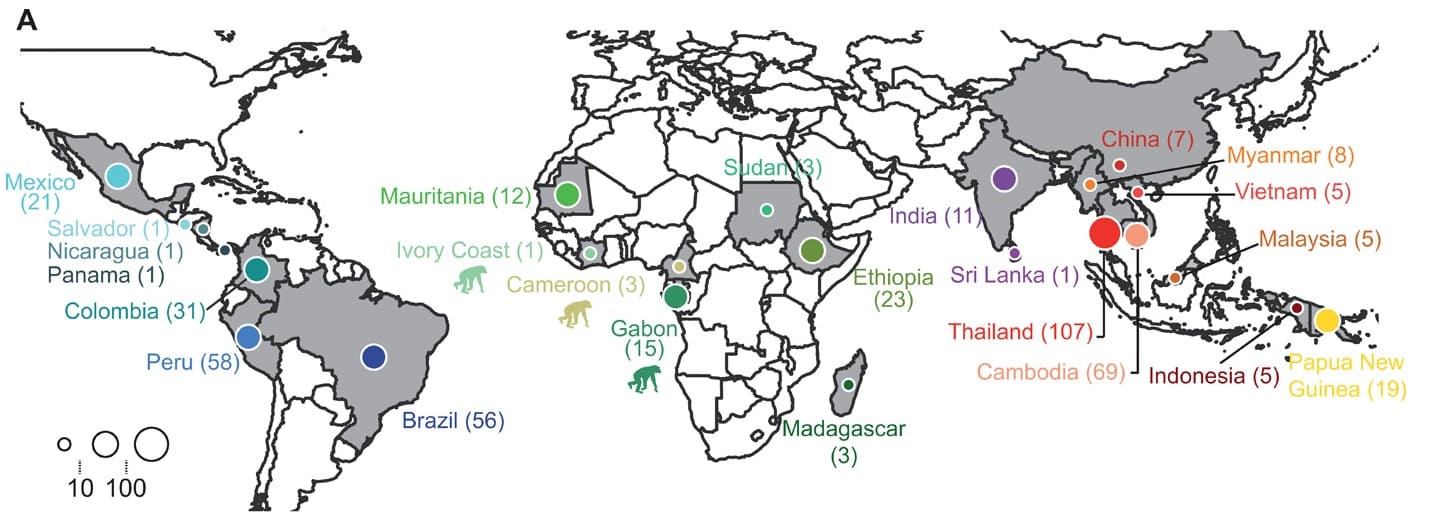
Studied sampling distribution
Figure legend:Country of origin of the 447 P. vivax and 19 P. vivax-like isolates used in the study of Draon J. et al. Within each country, isolates were collected at different locations. The chimpanzee pictogram represents African P. vivax-like isolates. @V.Rougeron
Main publications
https://doi.org/10.1371/journal.pntd.0008072
Portfolio
Photos legend : Field mission La Lekedi park, Gabon 2018, sanitary control – @V.Rougeron
Collaborators involved

Céline
ARNATHAU
Céline
ARNATHAU

Josquin
DARON
Josquin
DARON

Barthélémy
NGOUBANGOYE
Barthélémy
NGOUBANGOYE

Anne
BOISSIERE
Anne
BOISSIERE

Christine
SIDOBRE
Christine
SIDOBRE