research evolution adaptation plasmodium vivax-like african great apes monkeys
Plasmodium vivax, one of the five species responsible of human malaria
To unravel the origin of P. vivax clades: an Asian or African origin?
We will determine if the current dispersion of P. vivax reflects modern human migration
Completed Project
Recent News
Contact
- REHABS International Research Lab South Afrrica
- rougeron.virginie@gmail.com
- 06 46 64 23 18
More information about the project

@ V.Rougeron
Abstract
Plasmodium vivax, one of the five species responsible of human malaria, is characterized by the larger worldwide distribution in inter-tropical regions. P. vivax was considered to not circulate in the majority of the African continent since it was considered to be unable to infect Duffy negative humans. Indeed, the Duffy antigen was thought to be the unique portal of entrance of the parasite into the red blood cells. This dogma completely changed in the last years with the increased description of P. vivax infections in several African countries in both Duffy positive and negative individuals. During its history, P. vivax parasites colonized and evolved in different primate host species, including humans (P. vivax) and Central African great apes (gorillas and chimpanzees) (P. vivax-like). Today, the exact sequence of events that link P. vivax parasites in humans and great apes and that describe the migration roads of human P. vivax all around the world still remains unclear and the available genetic data are insufficient to robustly test evolutionary scenarios, specifically because of the lack of genetic information from African P. vivax genomes. Moreover, several studies published genomes of human P. vivax, but only few genomes have been described from the African continent (4 from Madagascar, 1 from Mauritania and 23 from Ethiopia recently). Through a retrospective approach based on genomic data the goal of the DMOV project is to get a better understanding of the ancestral geographic expansion of P. vivax populations and to decipher its origin. The DEMOV project is structured into two objectives, as follow:
Obj1. To unravel the origin of P. vivax clades: an Asian or African origin? When and how did P. vivax emerge in human and great apes populations? We will identify the origin of human P. vivax.
Obj2. To trace the routes of human P. vivax migrations all around the world P. vivax colonized different human populations all over the world, but the routes taken by the parasites are still unknown. We will determine if the current dispersion of P. vivax reflects modern human migration dispersal patterns and its roads of expansion wince its original emergence.
Funding
PEPS INEE-ECOMOB DMOV 2019, “Deciphering the migration and origin of human and african great apes Plasmodium vivax”
Principal Investigator: Virginie Rougeron – 14,800€ ; 10 months ; Mars 2019 – December 2019.

Partners



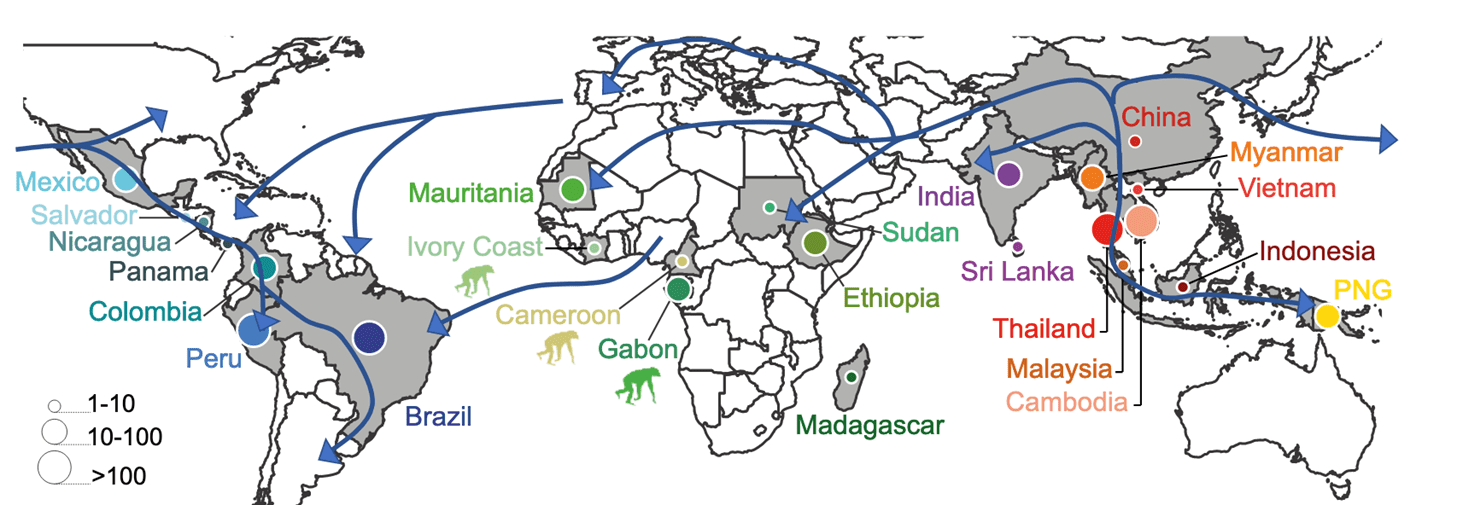
Studied sampling distribution
Distribution of the 1,154 genomes of P. vivax and the 37 genomes of P. vivax-like genomes analysed in this project. The pictograms represent the host species for P.vivax-like. The blue arrows represent the migration routes that will be migration routes that will be tested in this project, assuming an Asian origin of P. vivax.
Main publications
Portfolio
Landscapes in Gabon and Borneo. @ V. Rougeron
Collaborators involved

Céline
ARNATHAU
Céline
ARNATHAU

Josquin
DARON
Josquin
DARON

Franck
PRUGNOLLE
Franck
PRUGNOLLE