Population genomic evidence of Plasmodium vivax Southeast Asian origin
Abstract
Plasmodium vivax is the most common and widespread human malaria parasite. It was recently proposed that P. vivax originates from sub-Saharan Africa based on the circulation of its closest genetic relatives (P. vivax-like) among African great apes. However, the limited number of genetic markers and samples investigated questions the robustness of this hypothesis. Here, we extensively characterized the genomic variations of 447 human P. vivax strains and 19 ape P. vivax-like strains collected worldwide. Phylogenetic relationships between human and ape Plasmodium strains revealed that P. vivax is a sister clade of P. vivax-like, not included within the radiation of P. vivax-like. By investigating various aspects of P. vivax genetic variation, we identified several notable geographical patterns in summary statistics in function of the increasing geographic distance from Southeast Asia, suggesting that P. vivax may have derived from a single area in Asia through serial founder effects.
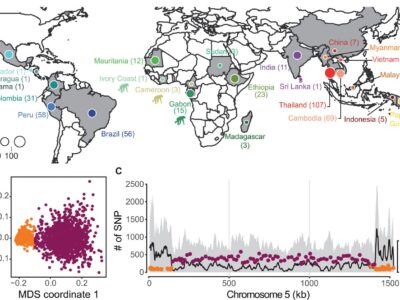
Fig. 1 Geographical origin of Plasmodium isolates and patterns of genomic variation.
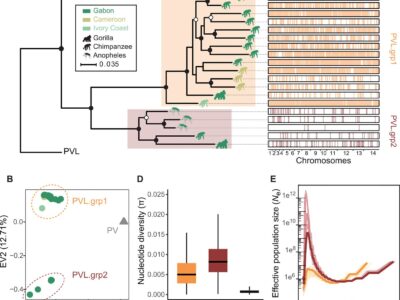
Fig. 2 P. vivax-like strains are structured in two distinct clades that form a sister monophyletic lineage to the human P. vivax.
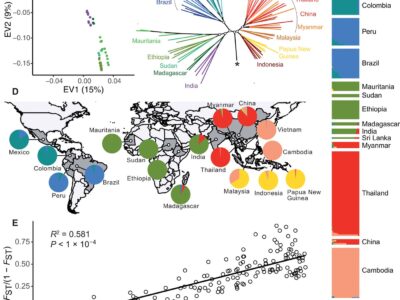
Fig. 3 The structure of P. vivax core region of the genome is mostly due to genetic isolation of natural populations.
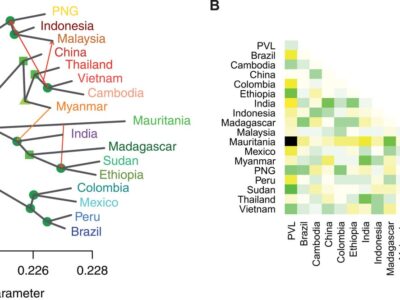
Fig. 4 Relationships and gene flow between P. vivax populations.
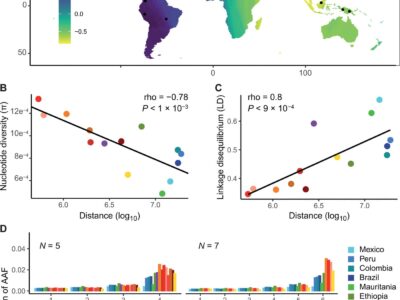
Fig. 5 The Southeast Asian origin of P. vivax is supported by patterns of nucleotide diversity, LD, and AAF.
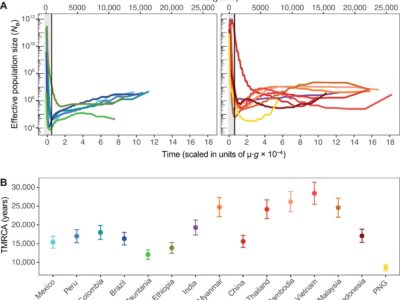
Fig. 6 Coalescent-based inference of the demographic history in each P. vivax population.

Publication infos
- Josquin Daron - Anne Boissière - Larson Boundenga - Barthelemy Ngoubangoye - Sandrine Houze - Celine Arnathau - Christine Sidobre - Jean-François Trape - Patrick Durand - François Renaud - Michael C. Fontaine - Franck Prugnolle - Virginie Rougeron
- josquin.daron@ird.fr - virginie.rougeron@cnrs.fr - virginie.rougeron@gmail.com
- Laboratoire MIVEGEC (Université de Montpellier-CNRS-IRD), 34394 Montpellier, France. • Centre of Research in Ecology and Evolution of Diseases (CREES), Montpellier, France. • Centre Interdisciplinaire de Recherches Médicales de Franceville, Franceville, Gabon. • Service de Parasitologie-mycologie CNR du Paludisme, AP-HP Hôpital Bichat, 46 rue H. Huchard, 75877 Paris Cedex 18, France. • Groningen Institute for Evolutionary Life Sciences (GELIFES), University of Groningen, PO Box 11103 CC, Groningen, Netherlands.
- This study was supported by ANR T-ERC EVAD, ANR JCJC GENAD, PEPS ECOMOB MOV 2019, CNRS, and MGX Montpellier sequencing facility. J.D. was supported by the Fondation pour la recherche Médicale (FRM, ARF20170938823) as well as by the Marie-Curie EU Horizon 2020 Marie-Skłodowska-Curie research and innovation program grant METHYVIREVOL (contract number 800489).
See the Publication
Science Advances 28 Apr 2021:
Vol. 7, no. 18, eabc3713
DOI: 10.1126/sciadv.abc3713